Welcome to the STimage-1K4M Dataset repository. This dataset is designed to foster research in the field of spatial transcriptomics, combining high-resolution histopathology images with detailed gene expression data.
Dataset Description
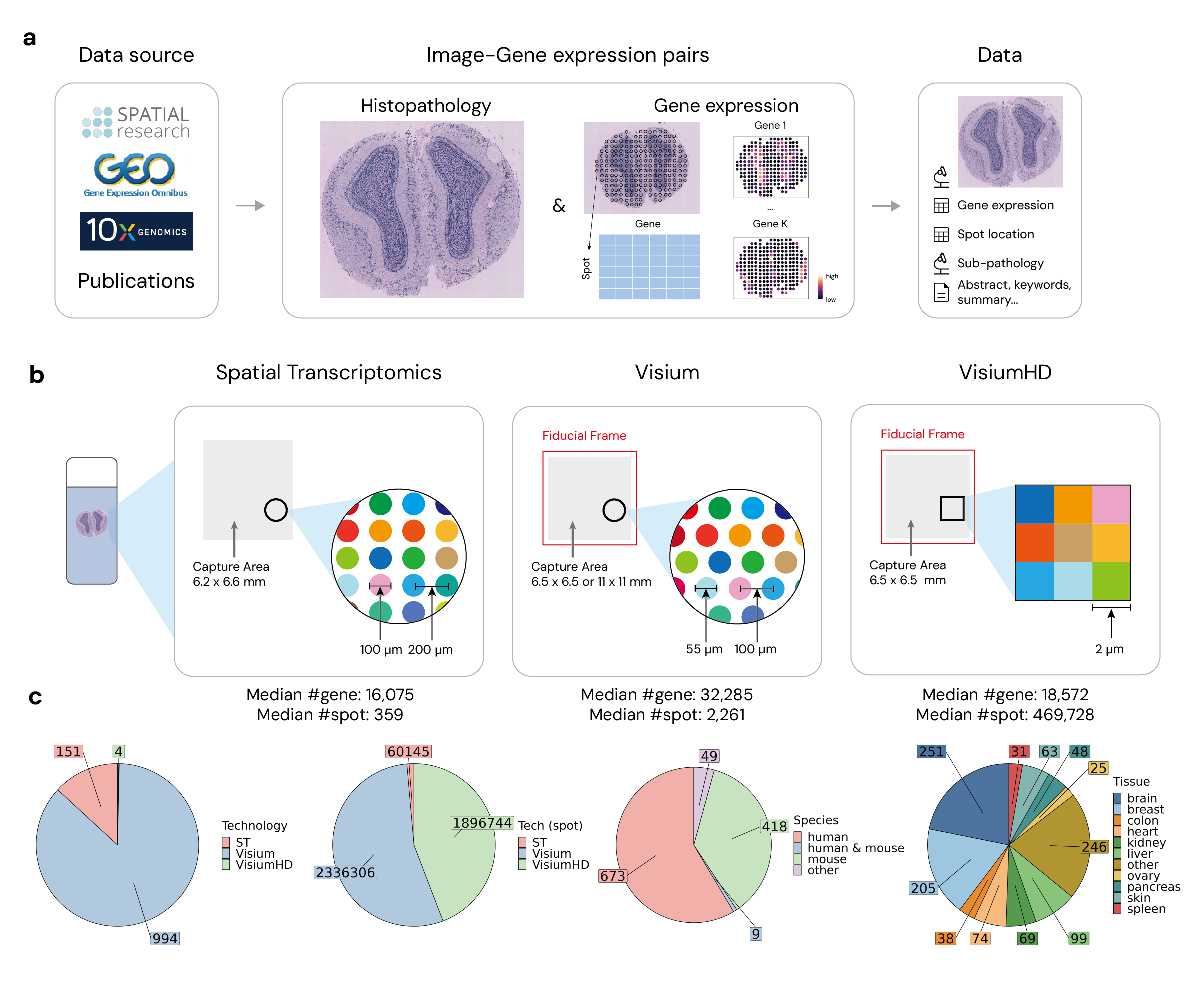
STimage-1K4M consists of 1,149 spatial transcriptomics slides, totaling over 4 million spots with paired gene expression data. This dataset includes:
- Images.
- Gene expression profiles matched with high-resolution histopathology images.
- Spatial coordinates for each spot.
See example folder for an example slide from Andersson et al. (pmid: 34650042).
Getting Started
To use the STimage-1K4M dataset in your research, please access the dataset via Hugging Face.
Data structure
The data structure is organized as follows:
├── annotation # Pathologist annotation
├── meta # Test files (alternatively `spec` or `tests`)
│ ├── bib.txt # the bibtex for all studies with pmid included in the dataset
│ ├── meta_all_gene.csv # The meta information
├── ST # Include all data for tech: Spatial Transcriptomics
│ ├── coord # Include the spot coordinates & spot radius of each slide
│ ├── gene_exp # Include the gene expression of each slide
│ └── image # Include the image each slide
├── Visium # Include all data for tech: Visium, same structure as ST
├── VisiumHD # Include all data for tech: VisiumHD, same structure as ST
Document
The code for data processing and reproducing evaluation result in the paper are in Document.
Acknowledgement
The fine-tuning and evaluation codes borrows heavily from CLIP and PLIP.
Citation
@misc{chen2024stimage1k4m,
title={STimage-1K4M: A histopathology image-gene expression dataset for spatial transcriptomics},
author={Jiawen Chen and Muqing Zhou and Wenrong Wu and Jinwei Zhang and Yun Li and Didong Li},
year={2024},
eprint={2406.06393},
archivePrefix={arXiv},
primaryClass={cs.CV}
}
License
All code is licensed under the MIT License - see the LICENSE.md file for details.
