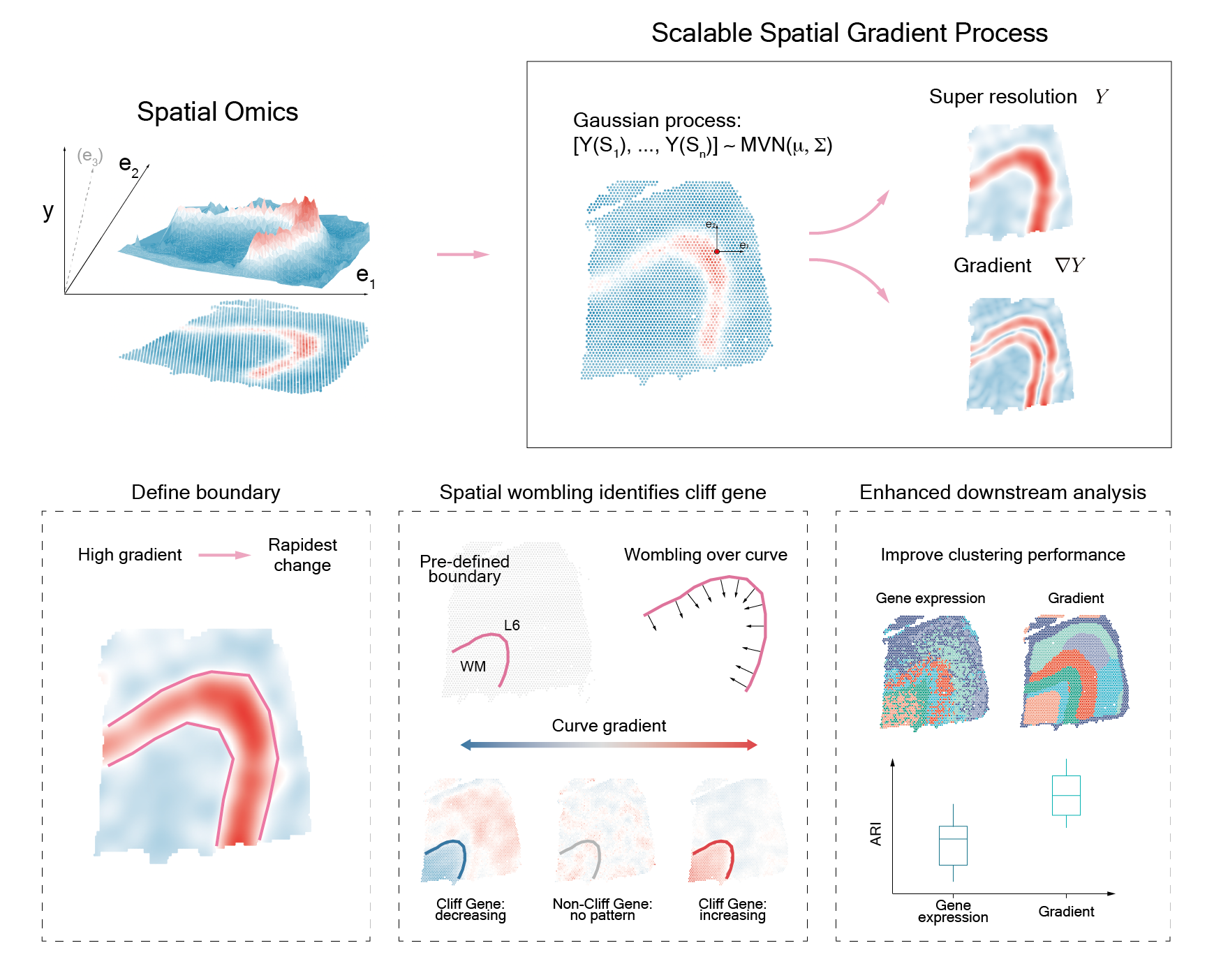
We present StarTrail, a novel gradient based method that powerfully defines rapidly changing regions and detects cliff genes, genes exhibiting drastic expression changes at highly localized or disjoint boundaries. StarTrail, the first to leverage spatial gradients for spatial omics data, also quantifies directional dynamics. Across multiple datasets, StarTrail accurately delineates boundaries (e.g., brain layers, tumor-immune boundaries), and detects cliff genes that may regulate molecular crosstalk at these biologically relevant boundaries but are missed by existing methods.
Features
- Boundary detection: Given spatial resolved feature (e.g. gene expression), StarTrail could detect gene expression boundary.
- Cliff gene detection: Given boundary, StarTrail could detect gene expressed stark change across the boundary.
- Downstream analysis enhancement: StarTrail could enhance downstream analysis by including gradient information in the task.
Usage
Following the instruction in Documents to use StarTrail.
Citation
Chen J, Xiong C, Sun Q, Wang GW, Gupta GP, Halder A, Li Y, Li D. Investigating spatial dynamics in spatial omics data with StarTrail. bioRxiv. 2024:2024-05.
@article{chen2024investigating,
title={Investigating spatial dynamics in spatial omics data with StarTrail},
author={Chen, Jiawen and Xiong, Caiwei and Sun, Quan and Wang, Geoffery W and Gupta, Gaorav P and Halder, Aritra and Li, Yun and Li, Didong},
journal={bioRxiv},
pages={2024--05},
year={2024},
publisher={Cold Spring Harbor Laboratory}
}
Contact
Please contact jiawenn@email.unc.edu or open an issue on GitHub for questions/comments