Highlights
Downloads (as of Jan 29th 2025): 100,852
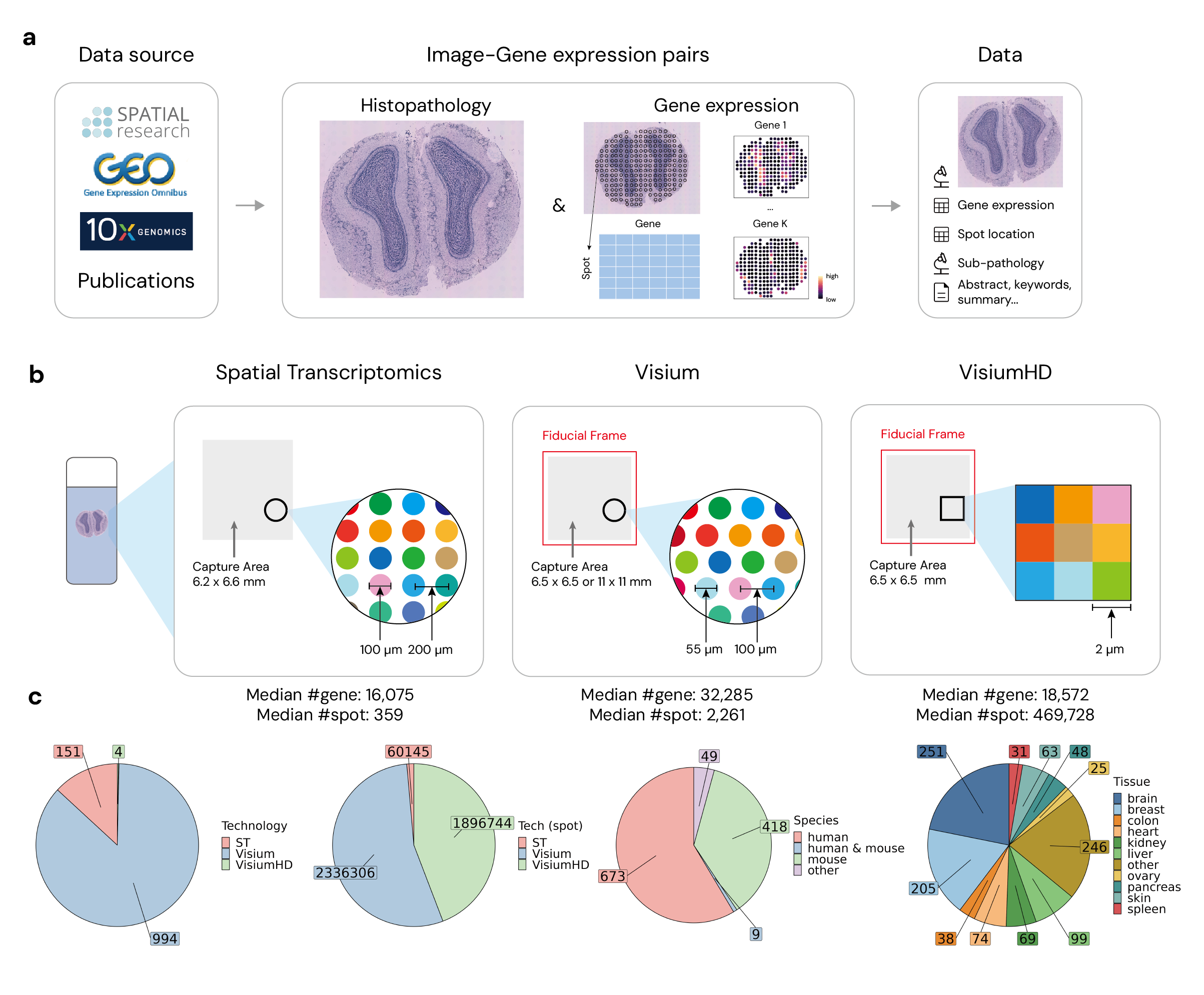
Recent advances in multi-modal algorithms have driven and been driven by the increasing availability of large image-text datasets, leading to significant strides in various fields, including computational pathology. In this study, we introduce STimage-1K4M, a novel dataset containing 1,149 images derived from spatial transcriptomics data, which captures gene expression information at the level of individual spatial spots within a pathology image. Specifically, each image in the dataset is broken down into smaller sub-image tiles, with each tile paired with 15,000-30,000 dimensional gene expressions. With 4,293,195 pairs of sub-tile images and gene expressions, STimage-1K4M offers unprecedented granularity, paving the way for a wide range of advanced research in multi-modal data analysis an innovative applications in computational pathology, and beyond.
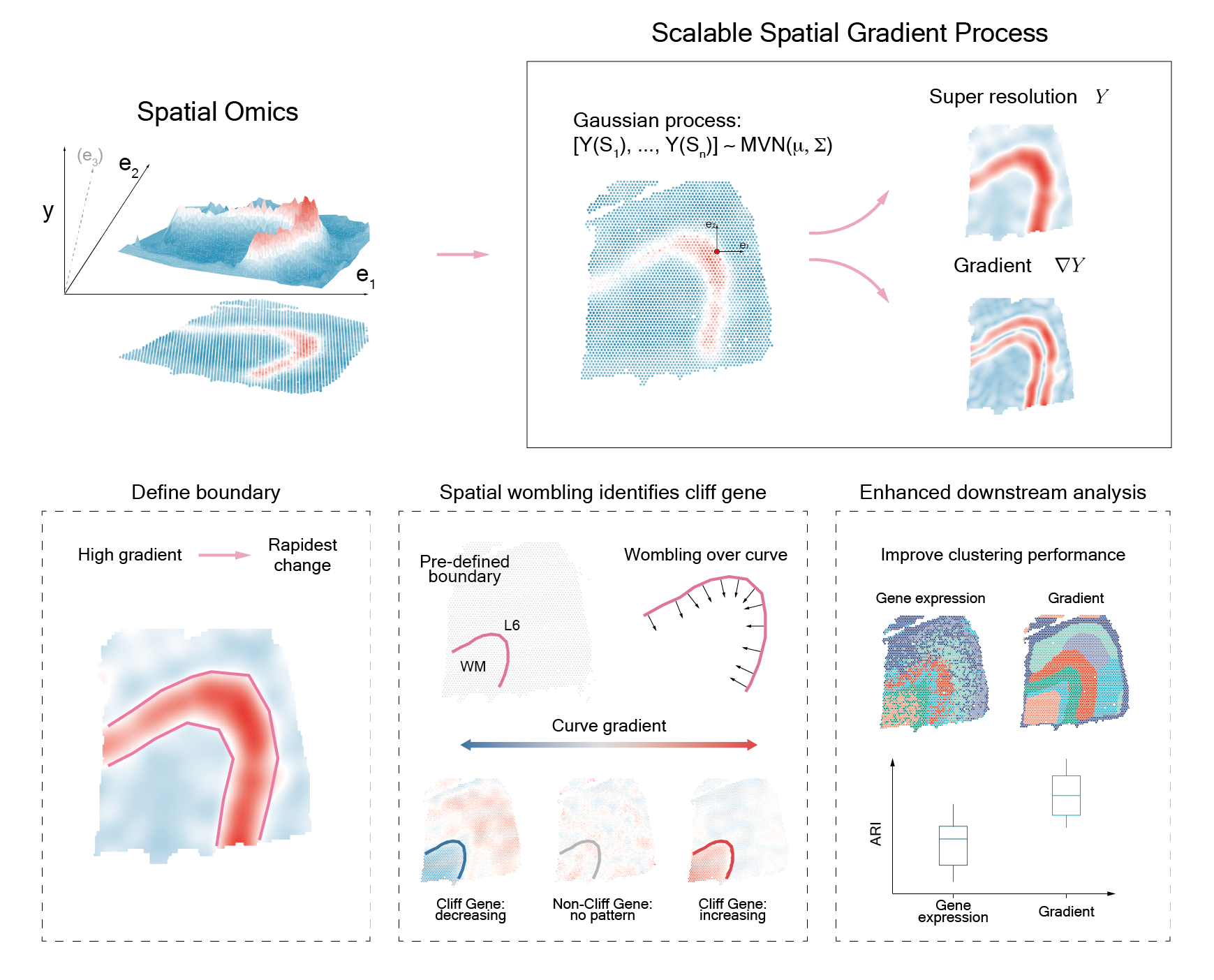
Spatial omics technologies revolutionize our view of biological processes. However, existing methods fail to capture localized, sharp changes characteristic of critical events (e.g. tumor development). Here, we present StarTrail, the first to leverage spatial gradients that powerfully defines rapidly changing regions, quantifies directional dynamics, and detects "cliff genes", genes exhibiting drastic expression changes at highly localized or disjoint boundaries. Across multiple datasets, StarTrail accurately delineates boundaries (e.g., brain layers, tumor-immune boundaries), and detects cliff genes that may regulate molecular crosstalk at these biologically relevant boundaries.
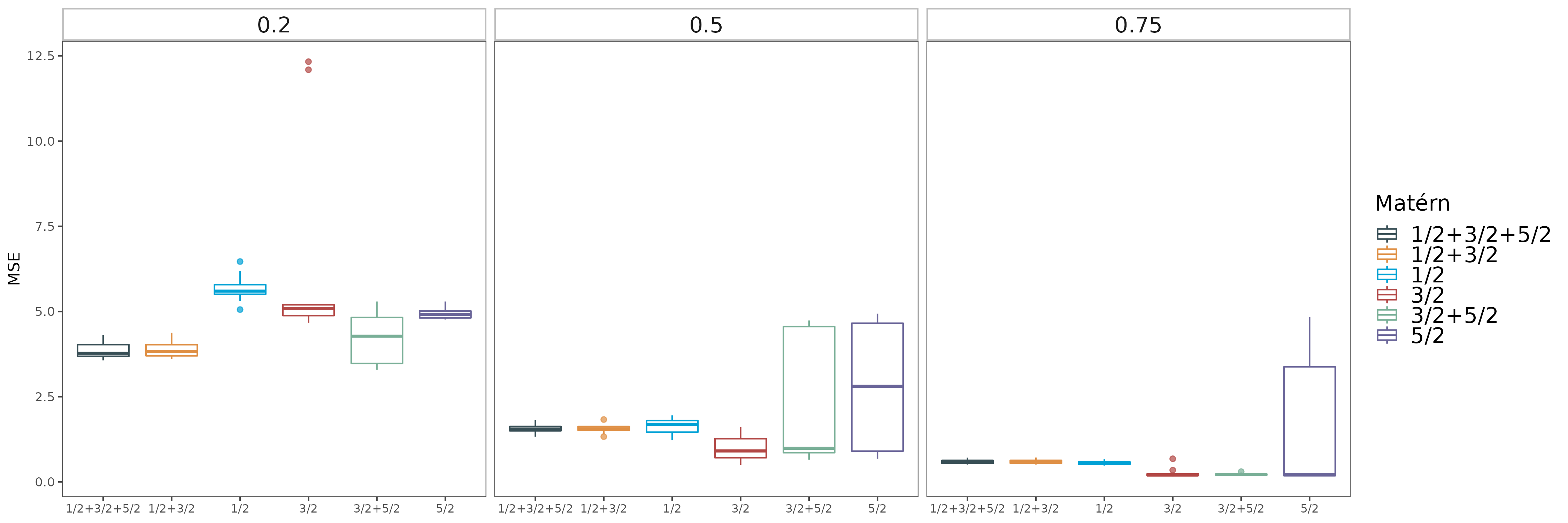
In this paper, we critically examine the prevalent practice of using additive mixtures of Matérn kernels in single-output Gaussian process (GP) models and explore the properties of multiplicative mixtures of Matérn kernels for multi-output GP models.
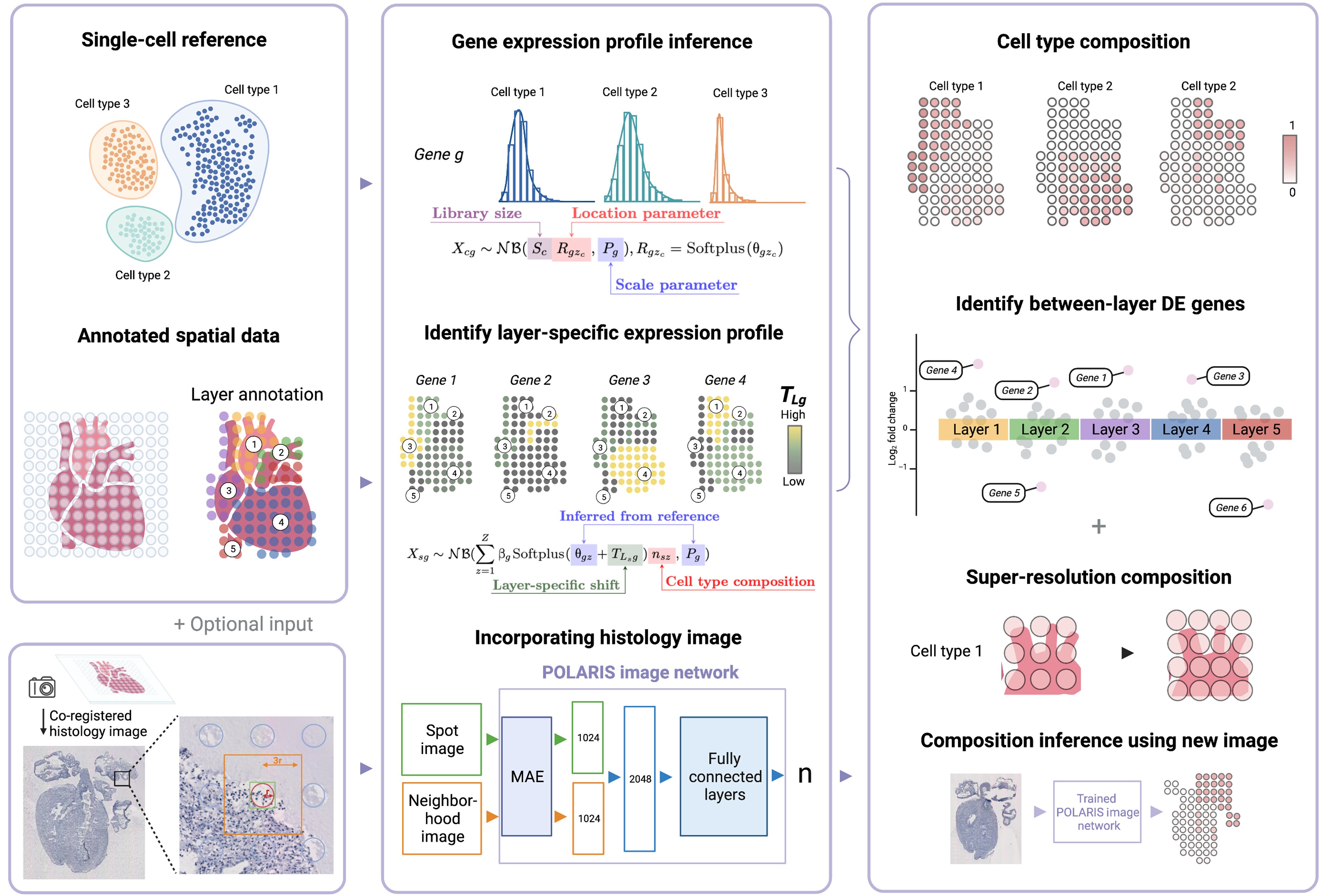
We present POLARIS, a versatile ST analysis method that can perform cell type deconvolution, identify anatomical or functional layer-wise differentially expressed (LDE) genes, and enable cell composition inference from histology images. Applied to four tissues, POLARIS demonstrates high deconvolution accuracy, accurately predicts cell composition solely from images, and identifies LDE genes that are biologically relevant and meaningful.
Full list of publication
\(^*\) co-first authorship, \(^\dagger\) co-last authorship, Paper Highlighted publication
Pre-prints & Submitted
- Xiong, C., Huang, S., Zhou, M., ..., Yao, H., Chen, J.$^\dagger$, Li, Y.$^\dagger$ (2025). A comprehensive comparison on clustering methods for multi-slide spatially resolved transcriptomics data analysis. bioRxiv, 2025-01.
- Chen, J., Halder, A., Li, Y., Li, D. (2024). Nearest-Neighbor Derivative Process. (Submitted to the Journal of the American Statistical Association (JASA)).
- Luo, T., Chen, J., Wu, W., Zhao, J., Yao, H., Zhu, H., & Li, Y. (2024). MAST-Decon: Smooth Cell-type Deconvolution Method for Spatial Transcriptomics Data. bioRxiv, 2024-05
- Jakubek, Y. A., Ma, X., Stilp, A. M., Yu, F., Bacon, J., Wong, J. W., ..., Chen, J., ... & Auer, P. L. (2024). Genomic and phenotypic correlates of mosaic loss of chromosome Y in blood. medRxiv, 2024-04. (Under review at The American Journal of Human Genetics (AJHG)).
- Chen, J., Xiong, C., Sun, Q., Wang, G. W., Gupta, G. P., Halder, A., Li, Y., Li, D. (2023). Investigating spatial dynamics in spatial omics data with StarTrail. bioRxiv, 2024-05. (Under revision at the Journal of the American Statistical Association (JASA)).
- Martin, C., Chen, J., Ye, A., Lodge, E., Ghastine, L., Dhingra, R., Hoyo, C. (2022). Differential methylation patterns in cord blood associated with gestational exposure to neighborhood crime: an epigenome-wide association study. (Revision submitted to Epigenetics).
- Wen, J., Li, G., Chen, J., Sun, Q., Liu, W., Guan, W., ... & Li, Y. (2022). DeepGWAS: Enhance GWAS Signals for Neuropsychiatric Disorders via Deep Neural Network. bioRxiv, 2022-12.
- Chen, J., You, J., Zhao, Z., Ni, Z., Huang, K., Wu, Y., ... & Lu, Q. (2020). Gamete simulation improves polygenic transmission disequilibrium analysis. bioRxiv, 2020-10. (Under revision at PloS Genetics). </ol> #### Published **Methodology**
- Mu, W.$^*$, Chen, J.$^*$, Davis, E. S., Reed, K., Phanstiel, D., Love, M. I., & Li, D. (2024). Gaussian Processes for Time Series with Lead-Lag Effects with Applications to Biology Data. arXiv preprint arXiv:2401.07400. (Accepted by Biometrics).
- Chen, J., Zhou, M., Wu, W., Zhang, J., Li, Y., & Li, D. (2024). STimage-1K4M: A histopathology image-gene expression dataset for spatial transcriptomics. arXiv preprint arXiv:2406.06393. In Thirty-eighth Conference on Neural Information Processing Systems (NeurIPS 2024).
- Sun, Q., Yang, Y., Rosen, J. D., Chen, J., Li, X., Guan, W., ... & Li, Y. (2024). MagicalRsq-X: A cross-cohort transferable genotype imputation quality metric. The American Journal of Human Genetics, 111(5), 990-995.
- Sun, Q., Rowland, B. T., Chen, J., Mikhaylova, A. V., Avery, C., Peters, U., ... & Li, Y. (2024). Improving polygenic risk prediction in admixed populations by explicitly modeling ancestral-differential effects via GAUDI. Nature communications, 15(1), 1016.
- Chen, J., Mu, W., Li, Y., & Li, D. (2023). On the Identifiability and Interpretability of Gaussian Process Models. In Thirty-seventh Conference on Neural Information Processing Systems (NeurIPS 2023).
- Lee, L., Yu, H., Jia, B. B., Jussila, A., Zhu, C., Chen, J., ... & Hu, M. (2023). SnapFISH: a computational pipeline to identify chromatin loops from multiplexed DNA FISH data. Nature Communications, 14(1), 4873.
- Jiang, M. Z., Aguet, F., Ardlie, K., Chen, J., Cornell, E., Cruz, D., ... & NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium, TOPMed Analysis Working Group. (2023). Canonical correlation analysis for multi-omics: Application to cross-cohort analysis. Plos Genetics, 19(5), e1010517.
- Chen, J., Luo, T., Jiang, M., Liu, J., Gupta, G. P., & Li, Y. (2023). Cell composition inference and identification of layer-specific spatial transcriptional profiles with POLARIS. Science Advances, 9(9), eadd9818.
- Rosen, J., Lee, L., Abnousi, A., Chen, J., Wen, J., Hu, M., & Li, Y. (2023). HPTAD: A computational method to identify topologically associating domains from HiChIP and PLAC-seq datasets. Computational and Structural Biotechnology Journal, 21, 931-939.
- Sun, Q., Yang, Y., Rosen, J. D., Jiang, M. Z., Chen, J., Liu, W., ... & Li, Y. (2022). MagicalRsq: Machine-learning-based genotype imputation quality calibration. The American Journal of Human Genetics, 109(11), 1986-1997.
- Chen, J.$^*$, Liu, W.$^*$, Luo, T.$^*$, Yu, Z., Jiang, M., Wen, J., ... & Li, Y. (2022). A comprehensive comparison on cell-type composition inference for spatial transcriptomics data. Briefings in Bioinformatics, 23(4), bbac245.
- Huang, L., Rosen, J. D., Sun, Q., Chen, J., Wheeler, M. M., Zhou, Y., ... & Li, Y. (2022). TOP-LD: A tool to explore linkage disequilibrium with TOPMed whole-genome sequence data. The American Journal of Human Genetics, 109(6), 1175-1181.
- Rosen, J. D., Yang, Y., Abnousi, A., Chen, J., Song, M., Jones, I. R., ... & Li, Y. (2021). HPRep: quantifying reproducibility in HiChIP and PLAC-seq datasets. Current Issues in Molecular Biology, 43(2), 1156-1170.
- Wu, Y., Zhong, X., Lin, Y., Zhao, Z., Chen, J., Zheng, B., ... & Lu, Q. (2021). Estimating genetic nurture with summary statistics of multigenerational genome-wide association studies. Proceedings of the National Academy of Sciences, 118(25), e2023184118.
- Huang, K., Wu, Y., Shin, J., Zheng, Y., Siahpirani, A. F., Lin, Y., Ni, Z., Chen, J., ... & Lu, Q. (2021). Transcriptome-wide transmission disequilibrium analysis identifies novel risk genes for autism spectrum disorder. PLoS genetics, 17(2), e1009309.
- Ren, X., Yang, H., Nierenberg, J. L., Sun, Y., Chen, J., Beaman, C., ... & Shen, Y. (2023). High-throughput PRIME-editing screens identify functional DNA variants in the human genome. Molecular Cell, 83(24), 4633-4645.
- Jakubek, Y. A., Zhou, Y., Stilp, A., Bacon, J., Wong, J. W., Ozcan, Z., ..., Chen, J., ..., & Auer, P. L. (2023). Mosaic chromosomal alterations in blood across ancestries using whole-genome sequencing. Nature Genetics, 1-8.
- Sullivan, P. F., Meadows, J. R., Gazal, S., Phan, B. N., Li, X., Genereux, D. P., ..., Chen, J., ... & Lindblad-Toh, K. (2023). Leveraging base-pair mammalian constraint to understand genetic variation and human disease. Science, 380(6643), eabn2937.
- Zhong, W., Liu, W., Chen, J., Sun, Q., Hu, M., & Li, Y. (2022). Understanding the function of regulatory DNA interactions in the interpretation of non-coding GWAS variants. Frontiers in cell and developmental biology, 10, 957292.
- Liu, W., Zhong, W., Chen, J., Huang, B., Hu, M., & Li, Y. (2022). Understanding regulatory mechanisms of brain function and disease through 3D genome organization. Genes, 13(4), 586.
- Wen, J.$^*$, Lagler, T. M.$^*$, Sun, Q.$^*$, Yang, Y., Chen, J., Harigaya, Y., ... & Li, Y. (2022). Super interactive promoters provide insight into cell type-specific regulatory networks in blood lineage cell types. PLoS Genetics, 18(1), e1009984.
- Sun, Q., Crowley, C. A., Huang, L., Wen, J., Chen, J., Bao, E. L., ... & Li, Y. (2022). From GWAS variant to function: A study of $\sim$ 148,000 variants for blood cell traits. Human Genetics and Genomics Advances, 3(1).
- Wen, J., Xie, M., Rowland, B., Rosen, J. D., Sun, Q., Chen, J., ... & Li, Y. (2021). Transcriptome-Wide association study of blood cell traits in African ancestry and Hispanic/Latino populations. Genes, 12(7), 1049.